

Our group is interested in different aspects of Bioinformatics, Computational Biology and Systems Biology. Our goal is to obtain new biological knowledge with an "in-silico" approach which complements the "in-vivo" and "in-vitro" methodologies of Biology. This mainly involves mining the massive amounts of information stored in biological databases.
Besides our lines of scientific research, we also collaborate with experimental groups providing them with bioinformatics support for their specific needs, and participate in different teaching projects. A brief summary of our main working lines follows. You can find more information in the links below and in our list of publications.
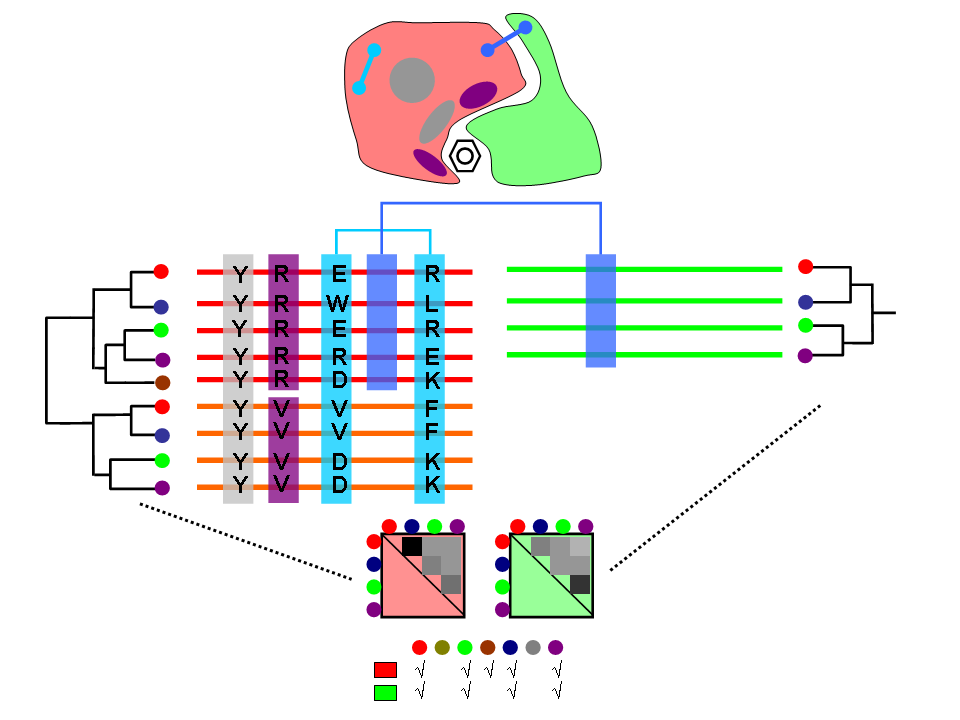
We have developed coevolutionary based approaches for predicting different protein features related to funcional sites and interactions.
Experimental determination of functional/active sites can not cope with the massive stream of new sequences comming from genome sequencing projects. Hence, computational methods for predicting sites with some functional importance in protein sequences and structures are highly demanded for this task. The methods we develop are based on the fact that functional sites are subject to certain evolutionary constraints whose landmarks can be detected on multiple sequence alignments.
The biological functions of many proteins can only be explained in the context of their relationshipts with others. Experimental techniques for the determination of interaction partners are still far from perfect and computational methods for predicting pairs of proteins which interact or are functionally associated have emerged. We have developed evolutionary-based methods for predicting interaction partners which have been accepted and followed by the community. These methods are mainly based on the hypothesis that interacting or functionally related proteins adapt to each other during the evolutionary process (co-evolution). We try to detect the landmarks that this co-evolutionary process left in the sequences and structures of the proteins.
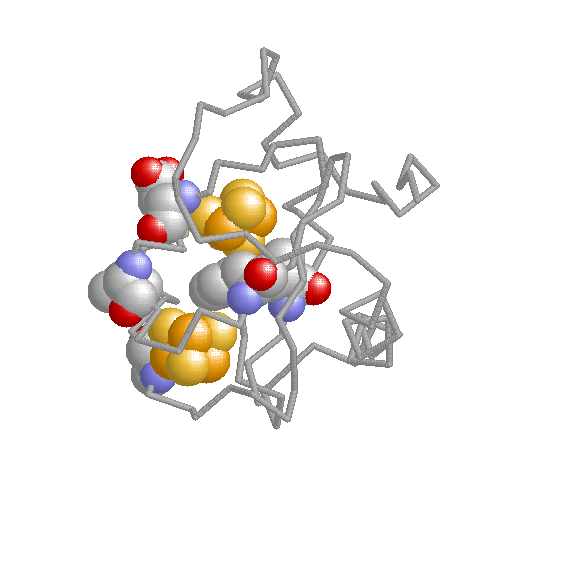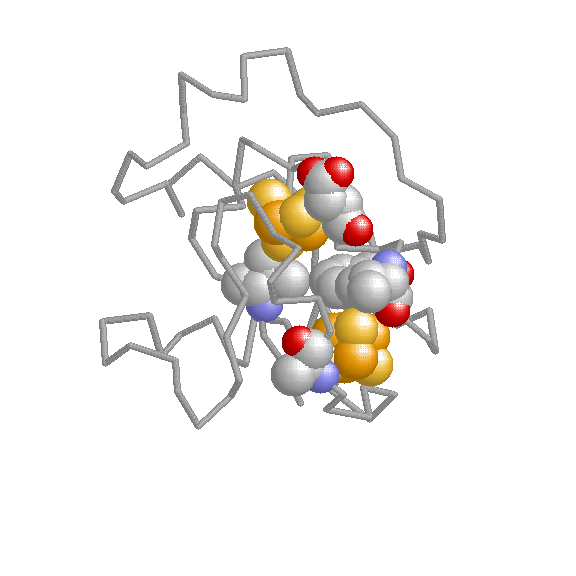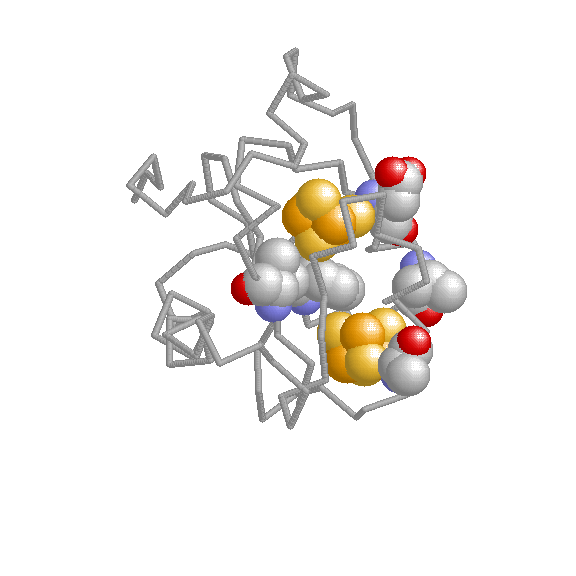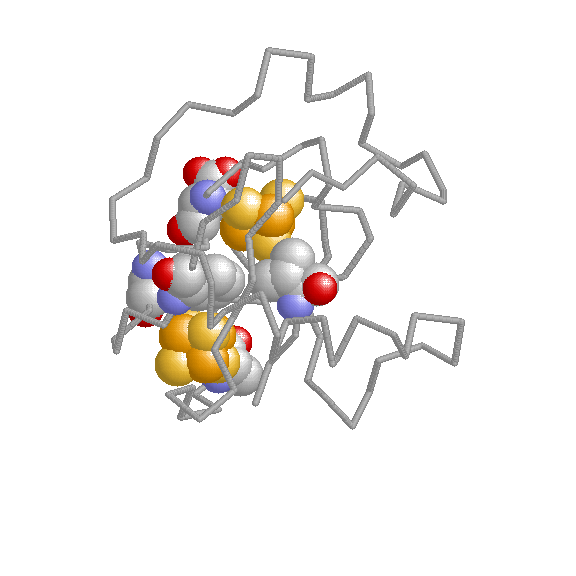
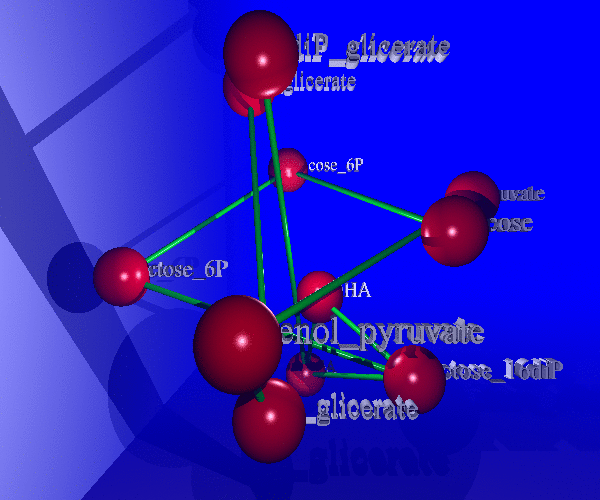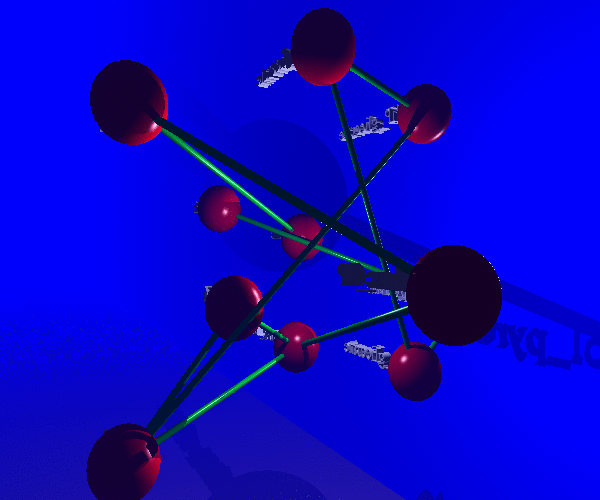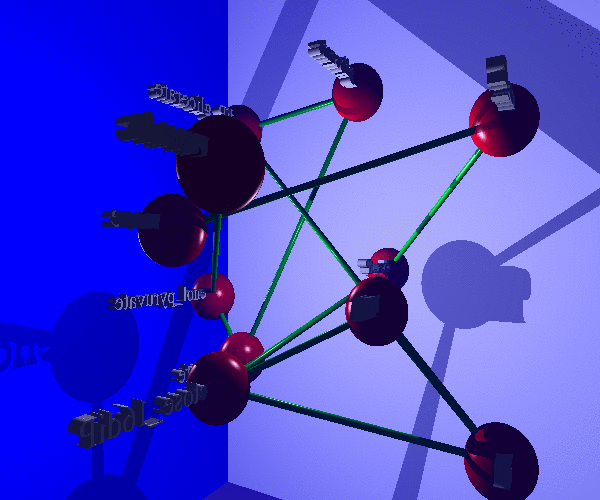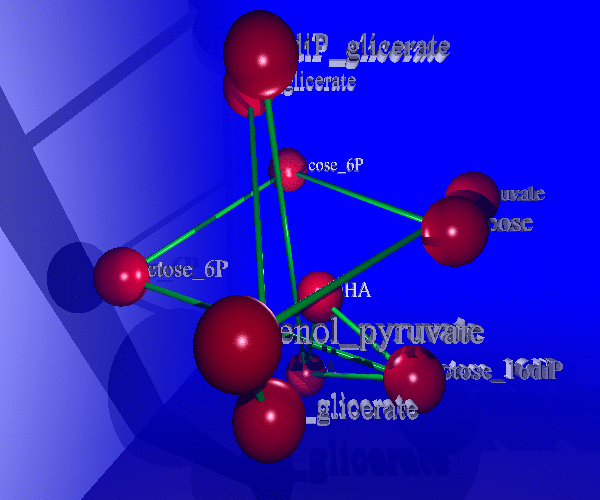
The study of living systems from a network perspective is providing new biological knowledge which could have never been obtained from the study of the individual components (genes, proteins, ...) no matter how detailed it was. As a prototype of complex systems, in biological systems many times "the whole is more than the sum of the parts". The biological knowledge obtained with this "top-down" approach is still modest compared with the whealt of information the "bottom-up" approach (exemplified by the Molecular Biology) has produced.
Nevertheless, it is clear that this new approach is required to complement the intrinsic limitations of the reductionist approach due to the complexity of living systems. We are studying metabolic networks (central metabolism and biodegradation) and protein interaction networks from this new "top-down" approach. Of special interest for us is the study of the complex phenomenon of "protein function" from a systemic perspective, trying to understand how complex functions arise by combining the molecular functions of proteins when these interact in intricate networks.
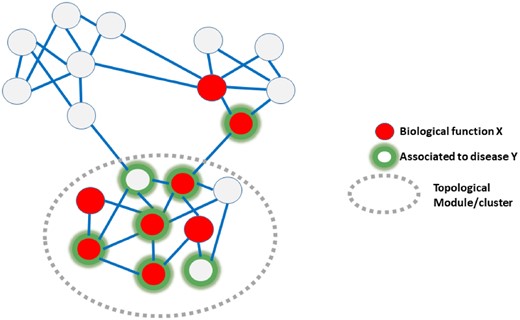
An active area of research in Biomedicine is the system-level study of cells through the analysis of molecular networks. In this framework diseases can be computationally study as a series of genetic and environmental perturbations that affect cellular functions. Molecular networks allow the integration and interpretation of multiple biological data (epigenomic, transcriptomic, proteomic, metabolomic) as well as clinical data. We are particularly interested in the systems-level study of clinical manifestations of rare diseases.
Most chemoinformatic approaches are still highly "molecule-centered" and lack the systemic and massive-data perspectives of bioinformatics. This paradigm is changing due to the accumulation of massive data on chemical compounds. We are interested in mining that increase ammount of data and explore the possibility of transposing bioinformatics approaches widely used in protein and DNA bioinformatics to chemical information.
Many research projects in Bioinformatics and Computational Biology have ended up in practical tools which can help in the daily work of experimental Molecular Biologists, specially in protein and DNA sequence analysis. In spite that many of these tools can be easily used by the community through web interfaces, others either are more difficult to use or their results are not straightforward to interpret without some knowledge on Bioinformatics. Moreover, many bioinformatics "protocols" are difficult to implement in automatic tools and they require manual intervention or expert knowledge to be carried out.
We provide experimental groups with bioinformatics expert knowledge, specially in the areas of protein sequence analysis (remote homology detection, domain identification, functional evolution, ...).
Related to this, we also have an academic interest in studying that large ecosystem of bioinformatic tools, quantify its characteristics, the life cycles of the tools, etc.