| |


- Files
- Control Panel
- Move & Rotate
- Display
- Rendering
- Tools
- Mutations
- Torsions
- Preferences
- hardware stereo
- electrostatics
- surface
- tips & tricks
Index







by N.Guex
& T.Schwede
|
|
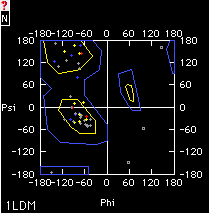
Ramachandran Plot
The Ramachandran Plot Window plots only values for the
currently selected amino-acids of the current layer. The name of the current
layer is drawn at the bottom left of the window. Amino-acids appear as
a little cross with the exception of Gly that appears as a square. To
know to which amino-acid a point belongs, simply place the mouse pointer
onto a point or a group of points. Its (their) name(s) will appear at
the top of the Window.
Omega, Phi, Psi and secondary structure assignment of
the currently displayed points can be saved as a tabulated text-file for
further analysis (Cmd + s on a Mac, Ctrl+s on a PC). Moreover, an image
can also be exported (Mac only). Core and allowed regions as defined by
Morris et al. ref [2] are also framed, and it is possible to select amino-acids
that are outside of these regions in one operation with the appropriate
item of the select menu.
Note: The Ramachandran plot can be used to directly modify a Phi/Psi
angle of a residue with a direct feedback of the modification. All you
need to do is drag a point at its new location. You can constrain the
rotation around either of the Phi or Psi axis by holding down the "9"
or "0" key while moving the point. By default it is the C-terminal part
of the protein that will move, as a little 'c' drawn just below the help
icon reminds you. If you want the N-terminal part to move, click on the
'c' that will be changed into a 'n'. If you want to move only a part of
the backbone (and not the whole backbone up to the C-terminal), you must
break the backbone between the last amino-acid you want to move and the
first you want to stay fixed. This is done with the appropriate item of
the tool menu.
|
 ExPASy Home page
ExPASy Home page ExPASy Home page
ExPASy Home page
 ExPASy Home page
ExPASy Home page