| |


- Files
- Control Panel
- Move & Rotate
- Display
- Rendering
- Tools
- Mutations
- Torsions
- Preferences
- hardware stereo
- electrostatics
- surface
- tips & tricks
Index







by N.Guex
& T.Schwede
|
|
User Guide
General Informations
Swiss-PdbViewer can load and display
several molecules simultaneously.
Each molecule is loaded into its own
layer. Each molecule is
composed of groups
(i.e. amino acids, nucleotides, substrates...). Each
group is composed of atoms, whose
coordinates are taken directly from a PDB file. Unless many
other packages, each time you move or rotate a molecule, the
actual coordinates of each atom is modified. When you save
back your molecule, its coordinates will be different unless
you reset its orientation or the "Save in Original
Orientation" item of the File Menu is enabled.
Workspace
The workspace is divided into several
windows
|

|
|
Here lays the main window,
where you can manipulate your molecules in
real time, measures distances and angles,
make mutations and so on...
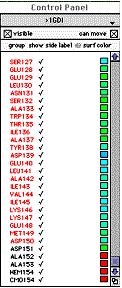
On the right
side, you've got a control Panel, that
provides a convenient way to select and
manipulate the attriubutes of individual
groups.
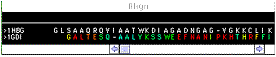
On the bottom
of your screen lays a window that shows
the alignment of your proteins, gives some
feedback informations and permit to thread
a sequence onto a reference in order to
submit an homology modelling request to
Swiss-Model.
|
|
|
|
|
|
 ExPASy Home page
ExPASy Home page ExPASy Home page
ExPASy Home page
 ExPASy Home page
ExPASy Home page