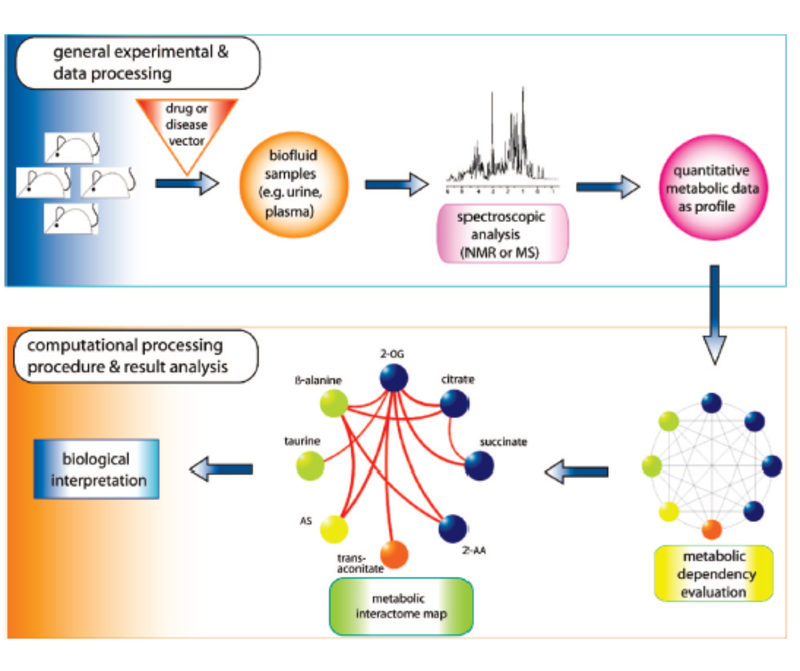
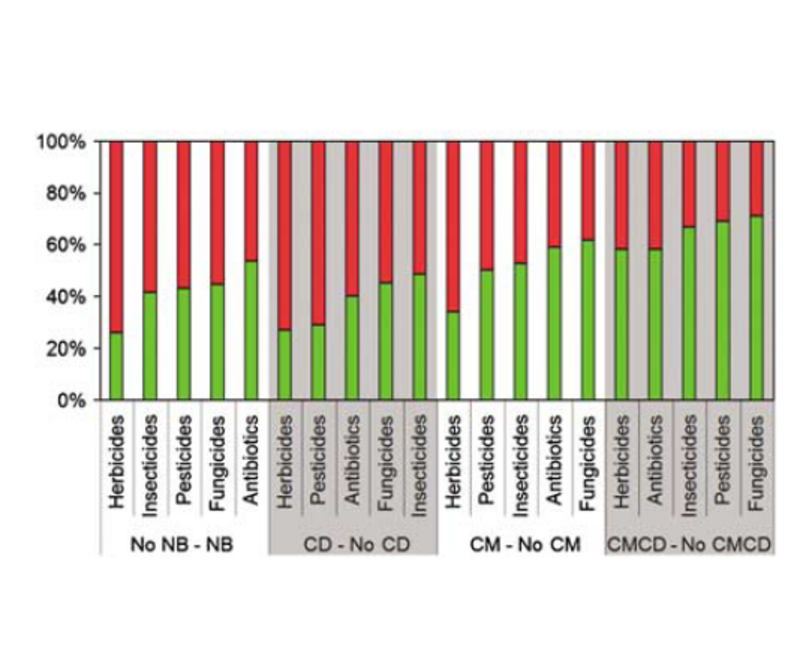
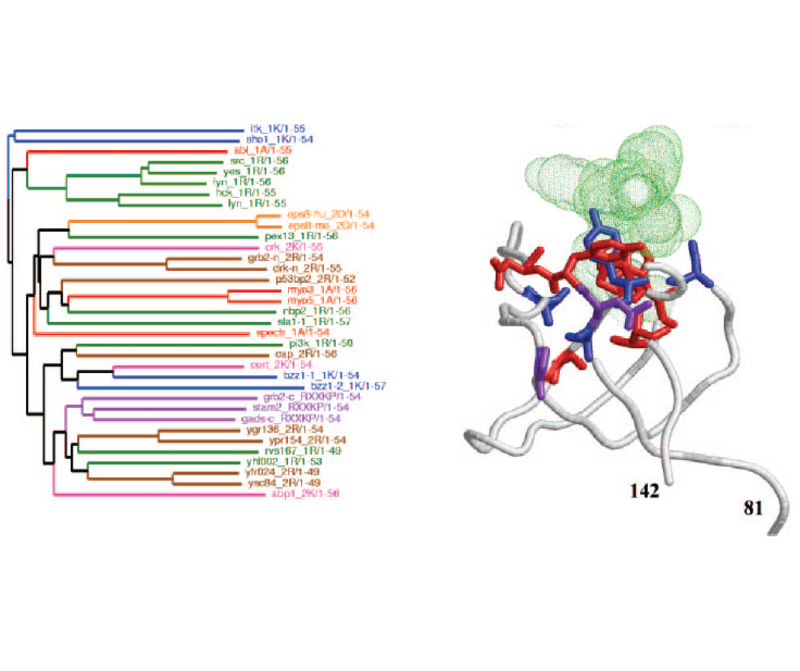
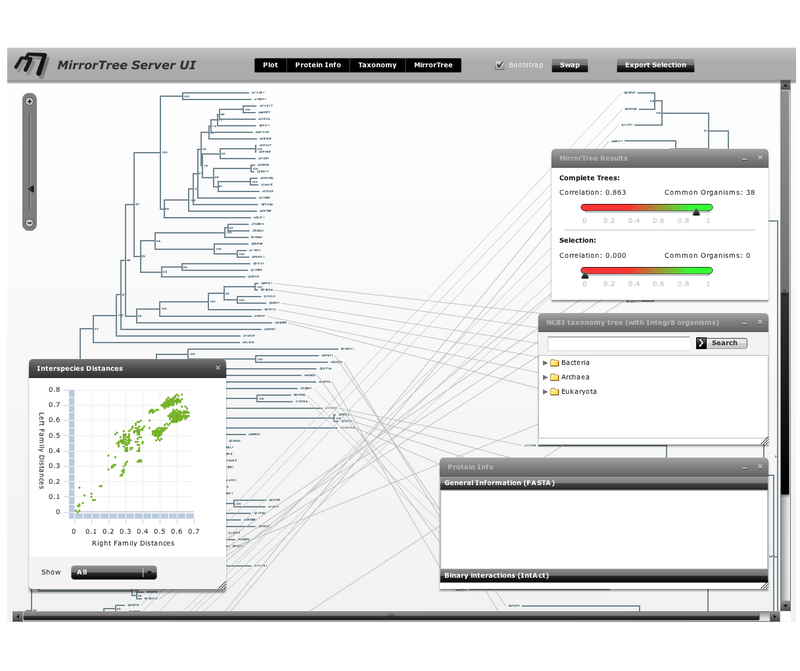
Welcome to the web space of the Computational Systems Biology Group. We are integrated in the Systems Biology Program of the National Centre for Biotechnology (CNB-CSIC). Our group is interested in different aspects of Bioinformatics, Computational Biology and Systems Biology. Our goal is to obtain new biological knowledge with an "in-silico" approach which complements the "in-vivo" and "in-vitro" methodologies of Biology. This mainly involves mining the massive amounts of information stored in biological databases.
Last updates
& changes
The web site was completely redesigned. If you have any comment/suggestion, please let us know.
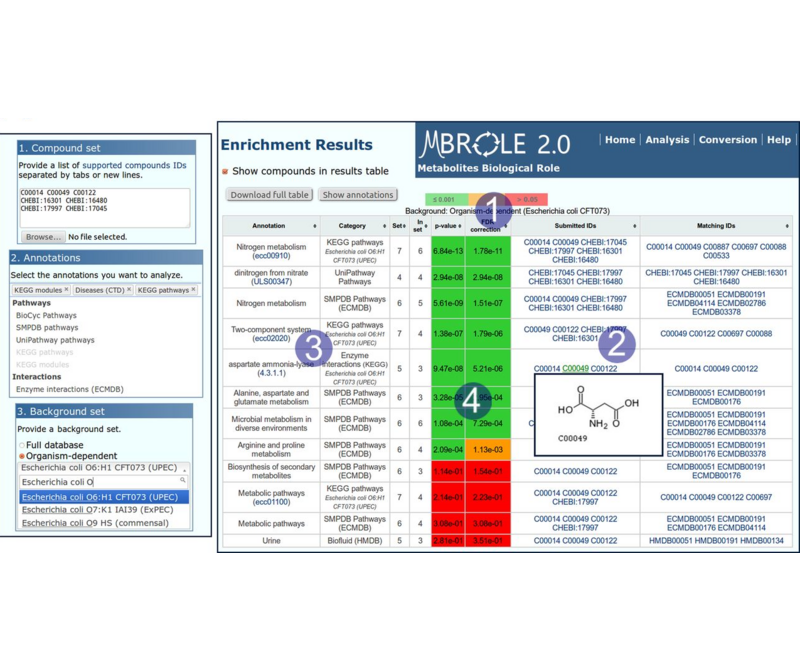
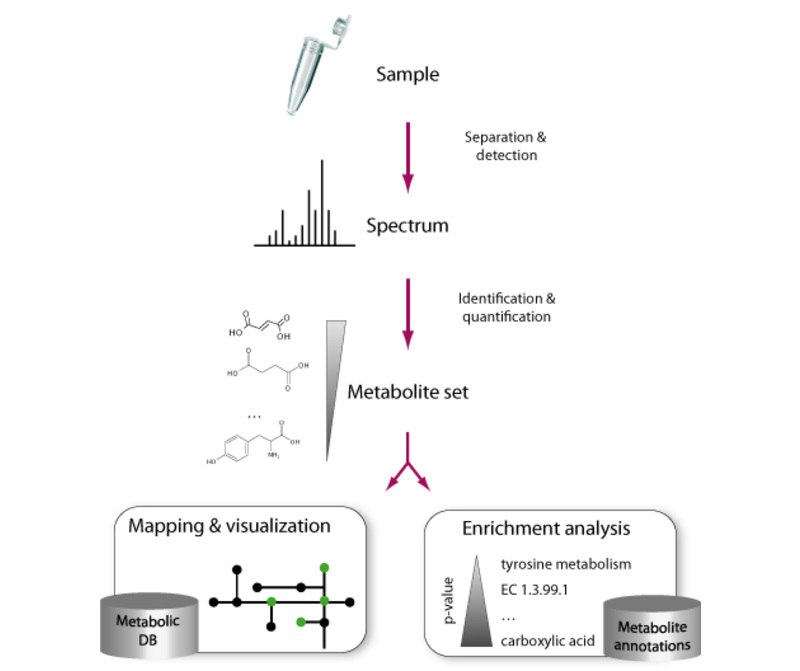
MBROLE 3.0 - Updated server for the functional analysis of metabolomics datasets. Lopez-Ibañez et al, 2023
CoMentG server for browsing millions of relationships between 9 types of biomedical concepts inferred from the scientific literature. (Novoa et al. 2024)
We have moved all web pages and services to a new web server. If you detect any problem, please let us know.
iFragMent server - Profile-based approach for predicting biological pathways of chemical compounds [Pubmed:34118870]