| sdsc1 | 3djigsaw | SwissModel | cphmodels | esypred |
1. sdsc1 | | 1.42 ± 2.02
[ 10] ( 2) | 1.73 ± 16.72
[ 1866] ( 1866) | 2.24 ± 13.66
[ 374] ( 374) | < 2 models | 2. 3djigsaw | -1.42 ± 2.02
[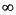 ] ( 2) ] ( 2) | | 0.17 ± 11.47
[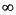 ] ( 4450) ] ( 4450) | -0.80 ± 14.48
[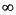 ] ( 114) ] ( 114) | 1.33 ± 11.91
[ 5824] ( 5824) | 3. SwissModel | -1.73 ± 16.72
[ 1866] ( 1866) | -0.17 ± 11.47
[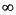 ] ( 4450) ] ( 4450) | | 1.86 ± 15.81
[ 1082] ( 1082) | 0.87 ± 12.19
[ 4907] ( 4907) | 4. cphmodels | -2.24 ± 13.66
[ 374] ( 374) | 0.80 ± 14.48
[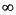 ] ( 114) ] ( 114) | -1.86 ± 15.81
[ 1082] ( 1082) | | -1.61 ± 13.55
[ 373] ( 373) | 5. esypred | < 2 models | -1.33 ± 11.91
[ 5824] ( 5824) | -0.87 ± 12.19
[ 4907] ( 4907) | 1.61 ± 13.55
[ 373] ( 373) | |