Super Secondary Structure
PART V
Helix-loop-helix motifs continued
A different helix-loop-helix motif is also common to certain DNA binding
proteins. This motif was first observed in prokaryotic DNA binding proteins
such as the cro repressor from phage lambda. This protein is a homodimer
with each subunit being 66 amino acids in length. Each subunit consists
of an all-antiparallel three stranded beta-sheet with three helical segments
inserted sequentially between the first and second beta-strands. The two
subunits of cro associate by virtue of the third beta-strands which interact
forming a six-stranded beta-sheet in the centre of the molecule. Mutagenesis
and biochemical work had indicated that residues in the second helix of
each cro monomer interacted with DNA. Accordingly model building studies
indicated that both these helices in the dimeric protein would fit into
the major groove of B-DNA. These proteins recognise base sequences which
are palindromic, i.e. possess an internal two-fold symmetry axis. The two
recognition helices of the cro protein are also related by a two-fold axis
passing through the central beta-sheet region of the dimer. Therefore,
the recognition helices of the cro dimer fit into the major groove of the
DNA and interact with each identical half of the palindrome. Hence, the
second helix of the helix-turn-helix motif has an important role in recognising
the DNA while the remainder of the structure serves to keep the two helices
in the correct relative position for fitting in the major groove of DNA.
Many other helix-turn-helix proteins with different folds exhibit essentially
the same mode of binding to DNA.
Beta-alpha-beta motifs
Antiparallel beta-strands can be linked by short lengths of polypeptide
forming beta-hairpin structures. In contrast, parallel beta-strands are
connected by longer regions of chain which cross the beta-sheet and frequently
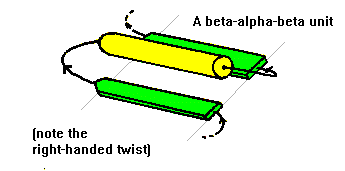
contain alpha-helical segments. This motif is called the beta-alpha-beta
motif and is found in most proteins that have a parallel beta-sheet. The
loop regions linking the strands to the helical segments can vary greatly
in length. The helix axis is roughly parallel with the beta-strands and
all three elements of secondary structure interact forming a hydrophobic
core. In certain proteins the loop linking the carboxy terminal end of
the first beta-strand to the amino terminal end of the helix is involved
in binding of ligands or substrates. The beta-alpha-beta motif almost always
has a right-handed fold as demonstrated in the figure.
The subsequent sections of this course on protein folds and domains
will demonstrate how the beta-alpha-beta motif is an important and widespread
element of supersecondary structure.
Recommended reading
Introduction to Protein Structure, C.Branden and J.Tooze, Garland,
New York (1991).
A novel super-secondary structure of proteins and the relation between
the structure and amino acid sequence, A.F.Efimov, FEBS Lett. 166,
33-38 (1984).
Structure of alpha-alpha-hairpins with short connections, A.V.Efimov,
Protein Engin.4, 245-250 (1991).
Structure of beta-beta-hairpins and beta-beta-corners, A.V.Efimov,
FEBS Lett., 284, 288-292 (1991).
Structure and evolution of calcium modulated proteins, R.H.Kretsinger,
CRC Crit.Rev.Biochem. 8, 119-174 (1980).
Comparison of the structures of cro and lambda repressor proteins
in bacteriophage lambda, D.H.Ohlendorf, W.F.Anderson, M.Lewis, C.O.Pabo
and B.W.Matthews, J.Molec.Biol. 169 757-769 (1983).
Conformation of beta-hairpins in protein structures, B.L.Sibanda,
T.L.Blundell and J.M.Thornton, J.Molec.Biol. 206, 759-777 (1989).
Beta-hairpin families in globular proteins, B.L.Sibanda and J.M.Thornton,
Nature 316, 170-174 (1985).
j.cooper 26/1/95
Last updated 7th April '97