 |
|
|
Membrane Proteins
The lipids are amphipathic , ie they have a hydrophilic polar 'head' and a hydrophobic non-polar 'tail', and they therefore spontaneously form bilayers in aqueous solution.
The membrane is fluid, with individual lipid molecules able to diffuse freely within the bilayer. It may therefore be thought of as a '2-dimensional solvent' for the proteins. Proteins diffuse 100 - 100,000 times slower than the lipids. There are three major types of lipids in membranes: phospholipids (the most abundant), glycolipids and cholesterol.
The composition of membranes varies depending on function. The plasma membranes of animal cells are approximately 50% protein in terms of weight. The inner mitochondrial membrane, which is involved in energy transduction, is about 75% protein, while the figure for myelin, which functions as an insulator, is roughly 25%. Many membrane proteins, termed glycoproteins, have covalently bonded carbohydrate groups. In mammalian cells such chains are often attached to an Asparagine's amide group. Carbohydrate chains occur on the extracellular side of plasma membranes, and the luminal side of intracellular membranes around organelles.
Useful links describing membrane structure:
![]() Mellitin 2mlt
(46Kb) [Bbk|BNL|ExP|Waw|Hal]
Use this short script to show the hydrophobic
and polar faces.
Mellitin 2mlt
(46Kb) [Bbk|BNL|ExP|Waw|Hal]
Use this short script to show the hydrophobic
and polar faces.
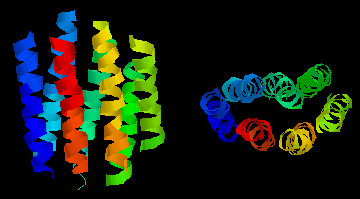 This diagram shows an example of such a "helix-bundle":
bacteriorhodopsin,
a light-driven proton-pump from the purple membrane of Halobacterium
halobium . The colours of the 7 helices are simply to distinguish them
in the two perpendicular views. This is a theoretical model.
This diagram shows an example of such a "helix-bundle":
bacteriorhodopsin,
a light-driven proton-pump from the purple membrane of Halobacterium
halobium . The colours of the 7 helices are simply to distinguish them
in the two perpendicular views. This is a theoretical model.
![]() Click here 1bac
(119Kb)[Bbk|BNL|ExP|Waw|Hal]
to examine the structure in more detail with RasMol. The 'backbone' or
'ribbons' options from the 'Display' menu best illustrate the arrangement.
The protein has a bound molecule of retinal which can be seen by selecting
'wireframe' mode, etc (select the 'chain' option in the 'Colours' menu).
Click here 1bac
(119Kb)[Bbk|BNL|ExP|Waw|Hal]
to examine the structure in more detail with RasMol. The 'backbone' or
'ribbons' options from the 'Display' menu best illustrate the arrangement.
The protein has a bound molecule of retinal which can be seen by selecting
'wireframe' mode, etc (select the 'chain' option in the 'Colours' menu).
![]() This rhodopsin
structure (Halobacterium halobium) was determined by electron diffraction
on two-dimensional crystals
1brd (124Kb)[Bbk|BNL|ExP|Waw|Hal]
to examine
This rhodopsin
structure (Halobacterium halobium) was determined by electron diffraction
on two-dimensional crystals
1brd (124Kb)[Bbk|BNL|ExP|Waw|Hal]
to examine
Here is another diagram of bacteriorhodopsin (by Manuel Peitsch at ExPASy). Also study the Bacteriorhodopsin Home Page (by PPS Consultant Iddo Friedberg of the Hebrew University, Jerusalem). If you are able to use MAGE, have a look at the Protein Science kinemages of bacteriorhodopsin modelling and refolding. There is a large super-family of receptors which adopt the same tertiary structure; to learn more about them try the G protein-Coupled Receptor DataBase.
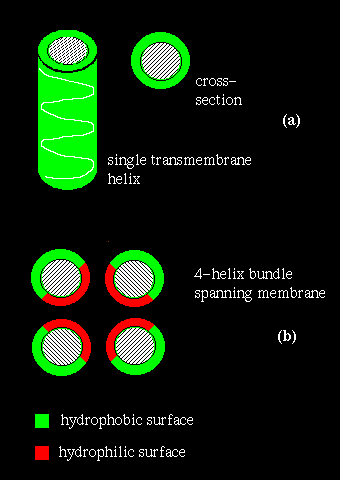
If several helices are bundled, then only the side chains on the outside of the bundle need be hydrophobic. This is illustrated in (b) above. If the central channel between the helices is lined with polar residues, the resulting structure might act as a pore in the membrane through which ions may pass.
Membrane proteins are difficult to crystallize. Because they have significant hydrophilic and hydrophobic surfaces, they are not soluble in aqueous buffer solutions yet they denature in organic solvents. Methods such as Infra-red Spectroscopy, Raman Spectroscopy and Circular Dichroism are used to deduce secondary structure.
However, other approaches may identify transmembrane regions. Naturally, one would expect a relatively long, uninterrupted sequence of hydrophobic residues to represent a membrane-spanning structure. On the other hand, a number of hydrophilic residues would be anticipated in a bundle structure such as the photosynthetic reaction centre (see also previous diagram (b). A quantitative measure of the hydrophobicity of a sequence is required.
Various hydrophobicity scales for amino acids based on for example
the free energy of transfer of the side chain from an inorganic solvent
to water have been compiled. For each position i in the sequence,
a
hydropathy index may be calculated as the mean hydrophobicity
value of all the residues from eg i-9 to i+9. Sharp peaks
in a plot of hydropathy index versus residue represent sequences which
would be unusually hydrophobic for a soluble protein and which therefore
are strong candidates for a transmembrane section: 
Such plots have in fact been found to be very successful in correctly predicting membrane-spanning sequences in structures which were subsequently elucidated by X-ray crystallography, such as the photosynthetic reaction centre.

A number of pigments (quinones) are bound between the helices of the photosynthetic reaction centre. Buried helix residues which interact either which the pigments, or with other helices are relatively highly conserved between different bacterial species, whereas those on the outside of the bundle are not. This indicates the non-specific nature of the hydrophobic interactions between the complex and the bilayer.
Click here for another diagram. Also look at the following (thanks to Manuel Peitsch at GLAXO Geneva):
It should be emphasized however that very few high-resolution crystal
structures of membrane proteins exist. Other examples to look at are:
![]() porin 3por (207Kb)[Bbk|BNL|ExP|Waw|Hal]
(from
Rhodobacter ) , which is composed mainly of beta-sheets in a 16-stranded
beta-barrel formation (see section on
all-beta
folds ) and forms a pore in the membrane 1.7 - 2.5 nm in diameter (shown
below); here are
1,
2
more images of porin.
porin 3por (207Kb)[Bbk|BNL|ExP|Waw|Hal]
(from
Rhodobacter ) , which is composed mainly of beta-sheets in a 16-stranded
beta-barrel formation (see section on
all-beta
folds ) and forms a pore in the membrane 1.7 - 2.5 nm in diameter (shown
below); here are
1,
2
more images of porin.
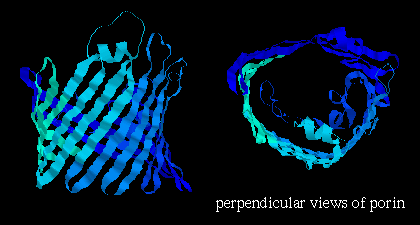
Note that the orientation of the strands is such that side chains alternately point into the interior and exterior of the pore; the former are strongly polar residues while the latter are very hydrophobic. Here is the Protein Science kinemage of the crystal structure. Click here for another kinemage.
![]() Also look at
the pore-forming domain of colicin A
1col
(260Kb) [Bbk|BNL|ExP|Waw|Hal].
(There are 2 of these domains in the asymmetric unit of this crystal structure).
Click
here
for a diagram. Colicin A is an antibiotic which is water-soluble, but which
can undergo a conformational change such that the pore-forming domain inserts
itself into the membrane of E. coli .
Also look at
the pore-forming domain of colicin A
1col
(260Kb) [Bbk|BNL|ExP|Waw|Hal].
(There are 2 of these domains in the asymmetric unit of this crystal structure).
Click
here
for a diagram. Colicin A is an antibiotic which is water-soluble, but which
can undergo a conformational change such that the pore-forming domain inserts
itself into the membrane of E. coli .
If you are able to use MAGE , click here for the Kinemage supplement to the Branden and Tooze "Introduction to Protein Structure" chapter on membrane proteins.
![]() Also look at the
two kinemages (1,
2)
of
Defensin 1dfn
(53Kb)[Bbk|BNL|ExP|Waw|Hal]
Also look at the
two kinemages (1,
2)
of
Defensin 1dfn
(53Kb)[Bbk|BNL|ExP|Waw|Hal]
 |
|
|
Last updated 7th April '97