Introduction
Nuclear receptors are a large family of structurally related ligand-inducible
transcription factors, including steroid receptors (SRs), thyroid/retinoids
receptors (TR, RARs and RXRs), vitamin D receptors (VDR), LXR, PPARs, estrogen
receptors (ERa and ERb),
and orphan receptors for which no ligand has been yet identified. While
having in common a modular structure, they are activated by distinct lipophilic
small molecules such as glucocorticoids, progesterone, estrogens, retinoids,
and fatty acid derivatives.
All nuclear receptors have a hydrophobic pocket into which its specific
ligand binds, with helix 12 (H12) being the key response element of NRís.
When an agonist is bound to a NR, H12 is oriented anti-parallel to H11,
capping the ligand binding pocket. This leaves a hydrophobic groove exposed
for the binding of coregulator proteins. When an antagonist is bound, H12
is displaced via an extended side chain. H12 moves outward, rotates, and
packs into the hydrophobic groove between helices 3, 4, and 5. As a result,
coactivators needed for transcription cannot bind. The following two images
show ERa with an agonist (left) and antagonist
(right) bound to it.
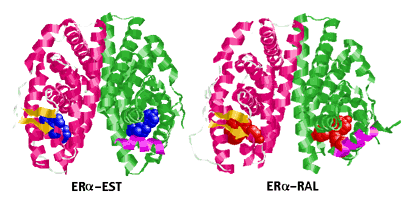 |
| Estrogen Receptor Ligand-Binding Domain Complexed
to Estradiol |
Estrogen Receptor Ligand-Binding Domain Complexed
to Raloxifene |
H12 is shown as a magenta coil in the "green sububits". The other subunit
in each structure is displayed as "Cartoons" with "Structure" coloring
(RasMol). The ligands are Spacefill.
Helix 12 Slide Show displays
JPEG views of several other ER-ligand complexes superimposed on ER-EST.
References to the structure papers are listed.
Two Different Estrogen Receptors
Recent studies have revealed the existence of two distinct estrogen
receptors in our bodies: ERa and ERb.
While they both bind estrogen as well as other agonists and antagonists,
the two receptors have distinctly different localizations and concentrations
within our body. Structural differences also exist between the two. allowing
for a wide range of diverse and complex processes to take place. The following
diagram, adapted from the Gustafsson review (1999), shows the distribution
of ERa and ERb

Structural Differences between ERa and
ERb
Two of the most interesting sites on the ER molecule are its ligand
binding domain (LBD), otherwise known as AF-2, and its growth factor binding
domain, otherwise known as AF-1. In addition, the DNA-binding domain (DBD)
is responsible for binding at estrogen response elements (ERE) on the chromosome.
A subtle difference between the two in their ligand-binding pockets is
the substituion of Leu 338 in ERa with Met 384
in ERb.
The following diagram, adapted from the Gustafsson review (1999), compares
the amino acid sequences of the two receptors:

The separate domains are identified in the ERa
diagram; the numbers in the ERb diagram show
the sequence identity as %.
Functional Differences
Interestingly, ERa and ERb,
when complexed with estrogen, were shown to signal in opposite ways from
an AP1 site, with estrogen activating transcription in the presence of
ERa and inhibiting transcription in the presence
of ERb. The ER ligands tamoxifen, raloxifene,
and ICI-164384 were activators with ERb as well
as ERa, although the degree of agonism differed
between cell types. These molecules are examples of SERMís, selective estrogen
receptor modulators. Thus, the role of estrogen complexed to ERb
appears to be to turn off transcription of these genes, whereas the SERMs
may override this blockade and activate gene transcription.
Effects of SERM's
Most recent drugs targeted to the ER, such as tamoxifen, ICI-164384,
and raloxifene act as either ER antagonists or agonists depending on the
species, tissue, and the dose administered. For instance, raloxifene has
been reported to act as an antiestrogen in breast tumor tissue and the
brain, while it has potentially beneficial estrogen-like effects in bone
and in modulating factors associated with cardio-vascular diseases. Another
example is tamoxifen, which was developed as an antiestrogen for the treatment
of breast cancer and was subsequently shown to have estrogen-like effects
on bone and the cardiovascular system. However, the potentially beneficial
effects of tamoxifen in reducing the risk of osteoporotic fractures and
coronary heart disease in postmenopausal women are at least partially offset
by its estrogenic effects on the uterus, increasing risk of endometrial
cancer development.
Binding Assays
Typical assays to gauge the binding affinities of estrogen analogs
or new drugs follow a standard pattern. The receptors are infused with
E2, and a binding curve is obtained based on varying concentrations of
E2 present. Then, the molecule in question is added in varying concentrations,
acting as a competitive inhibitor, and its ability to bind is plotted against
E2's. Most of these assays are depicted using Scatchard plots. The relative
binding constants of some well-studied molecules are as follows:
| Ligand |
ERa |
ERb |
| 17b-Estradiol ("E2"
or "EST") |
100 |
100 |
| Diethylstilbestrol |
468 |
295 |
| Tamoxifen |
6 |
7 |
The following chart compares the binding of reservatrol to that of estrogen.
The phytochemical resveratrol, which is found in grapes and wine, has been
reported to have a variety of anti-inflammatory, anti-platelet, and anti-carcinogenic
effects. Based on its structural similarity to diethylstilbestrol, a synthetic
estrogen, it was thought to be a phytoestrogen. The following figure
was taken from an article by Gehm, et al. (1997):

Estrogen receptor binding assays were performed by using 0.1 nM (circles),
0.3 nM (triangles), or 1.0 nM (squares). 125I-estradiol competed
with the indicated concentrations of resveratrol (open symbols) or unlabeled
estradiol (filled circles). Each point represents the mean and range of
duplicate assays after subtraction of nonspecific binding. All results
are shown as percentage of binding in the absence of competitor.
As can be seen from the graph, estradiol has a much higher binding affinity
for ER than does resveratrol. It is important to note that binding affninties
in vitro are often much higher than those observed in vivo due to the numerous
other pathways and cellular processes that may interfere with binding.
Coactivators and Corepressors
Transactivation requires the recruitment of coactivators, such as SRC-1,
that posess histone acetyltransferase activity or can recruit a histone
acetyl transferase. This complex can decompact the chromatin, enabling
a transcription initiation complex to form. Silencing involves the recruitment
of corepressors, such as SMRT, and histone deacetyltransferases.
The transconformation of H12, along with other associated structural
changes, creates a surface on the receptor that can bind coactivators such
as SRC-1. These coactivators contain one or more "LXXLL boxes" that are
responsible for nuclear receptor binding, where L is leucine and X is any
amino acid in the sequence motif.
Phytoestrogens
The phytoestrogen, genistein, is completely buried within the hydrophobic
core, but H12 does not adopt the distinctive "agonist" position. Instead,
H12 lies in a similar orientation to that observed with ER antagonists.
The suboptimal alignment of the transactivation helix results in genistein's
partial agonist character with ERb. The following
is the structure of ERb complexed to genistein
and raloxifene:
 |
| ERb Ligand-Binding
Domain Complexed to Genistein |
ERb Ligand-Binding
Domain Complexed to Raloxifene |
H12 is shown as a magenta coil in the "green sububits". The other subunit
in each structure is displayed as "Cartoons" with "Structure" coloring
(RasMol). The ligands are Spacefill.
Estrogen: It's Not Just for Females!
Estrogen has been shown to influence the morphology and function of
the secretory epithelial cells in rat prostates, making it likely that
at least some of the effects of estrogens in vivo are direct rather
than indirect. It is unclear whether the described effects are mediated
by ERa or ERb, or
perhaps even by both receptors, although the expression of ERb
mRNA and protein seems to be higher than that of ERa.
Another interesting fact is that that plasma estrogen levels increase with
age in men. Low levels of estrogen are one of the key factors in bringing
on osteoporosis, as estrogen helps maintain bone strength in both men and
women. The "Osteoporosis in Males" page, linked below explains this further.
Page Top
Selected References
These articles were either published recently, or of particular interest
for our reading, or both. Consult the reviews for more comprehensive listings.
Reviews
Gustafsson, J-Å (1999): "Estrogen receptor b
- a new dimension in estrogen mechanism of action". J Endocrinol
163:379-383.
**Figures 1 & 2 shown above.
Moras, D & Gronemeyer, H (1998) "The nuclear receptor ligand-binding
domain: structure and function". Curr Opin Cell Biol 10:384-391.
**This paper gives a good overview of the details surrounding silencing
and transactivation abilities of the estrogen receptor. Their explanations
are sequential and easy to follow. It clarifies much of the coactivator
and corepressor recruitment that is alluded to in many other papers regarding
this topic.
Pettersson, K & Gustafsson, J-Å (2001): "Role of estrogen
receptor beta in estrogen action". Ann Rev Physiol 63:165-192.
**Summary of molecular and physiological aspects of ER functions.
Glass, C & Rosenfeld, MG (2000): "The coregulator exchange in transcriptional
functions of nuclear receptors". Genes Dev 14:121-141.
**Extensive description of transcriptional coactivators and corepressors
in the larger context of the NR superfamily. A good starting point for
another Independent Reading project.
Simpson, ER & Davis SD (2000) "Another role highlighted for estrogens
in the male: Sexual behavior". PNAS 97:14308-14340.
**Short review and Commentary on a research paper in the same issue
of PNAS.
Research Papers
Pike, ACW, et al. (1999) "Structure of the ligand-binding domain of
estrogen beta in the presence of a partial agonist and a full antagonist".
EMBO J 18:4608-4618, 1999.
**This paper gives good insight into the intricacies of the ER in regards
to the minute comformational changes brought upon by binding of raloxifene
and genistein, and how these changes propagate to affect transcriptional
activity.
Gehm, BD, et al. (1997) "Resveratrol, a polyphenolic compound found
in grapes and wine, is an agonist for the estrogen receptor". PNAS
94:14138-14143.
**Figure 1 shown above.
Stauffer, SR, et al. (2000) "Pyrazole ligands: Structure-affinity/activity
relationships and etsrogen receptor-a-selective
agonists". J Med Chem 43:4934-4947.
**Synthesis and characterization of a compound with ~400-fold binding
preference for ERa. Binding measurements, transcription
assays, and structural modeling are reported.
Mak, HY, et al. (1999) "Molecular determinants of the estrogen receptor-coactivator
interface". Mol Cell Biol 19:3895-3903.
**Mutagenesis experiments show that more than the LXXLL motif is required
for specifc binding.
Gee, AC, et al. (1999) "Coactivator proteins have a differential stabilizing
effect on the binding of estrogens and antiestrogens with the estrogen
receptor". Mol Endocrinol1912-1923.
**A peptide corresponding to the SRC-1 NR Box 2 was used with a fluorescent
estrogen analog to measure binding to the protein complexes.
Thornton, JW (2001) "Evolution of vertebrate steroid receptors from
an ancestral estrogen receptor by ligand exploitation and serial expansions".
PNAS 98:5671-5676.
**Phylogenetic analysis used to propose the relationships between all
steroid receptors and a possible role for orphan receptors in vertebrate
evolution.
Page Top
Links to Related ER Sites
General Topics
NucleaRDB: An Information
System for Nuclear Receptors
http://receptors.ucsf.edu/NR/
Nuclear Receptor
Resource: Structures, Graphics, etc.
http://nrr.georgetown.edu/NRR/NRR.html
The Steroid Receptor
Associated Proteins Resource: at Medical College of Ohio
http://www.mco.edu/depts/pharm/srapr.html
Nuclear
Hormone Receptors
http://www.ks.uiuc.edu/Research/pro_DNA/ster_horm_rec/
Osteoporosis
in Males
http://uwcme.org/courses/bonephys/opmale.html
Environmental Estrogens
and Other Hormones (EEOH)
http://www.tmc.tulane.edu/ecme/eehome/
Chime Pages Elsewhere
Estrogen
Receptor (Steroid Binding Domain): Overview and part of a tutorial
on several proteins.
http://www.amherst.edu/~pbohara/biochem_30/chime/frameset-h.htm
Estrogen
Receptor (DNA Binding Domain)
http://www.amherst.edu/~pbohara/biochem_30/chime/frameset-i.htm
ER
LBD: Features of the Tanenbaum, et al. (1998) structure.
http://srv2.lycoming.edu/~newman/courses/bio43598/ER-ligand/index.html
ER
DBD
http://srv2.lycoming.edu/~newman/courses/bio43598/ER-DNA/index.html
Page Top |
 Estrogen Receptor Structures & Functions
Estrogen Receptor Structures & Functions



